by Matthew Cobb
There’s a great XKCD up today (click to embiggen, I’ve had to shrink it to not bump into the ads):
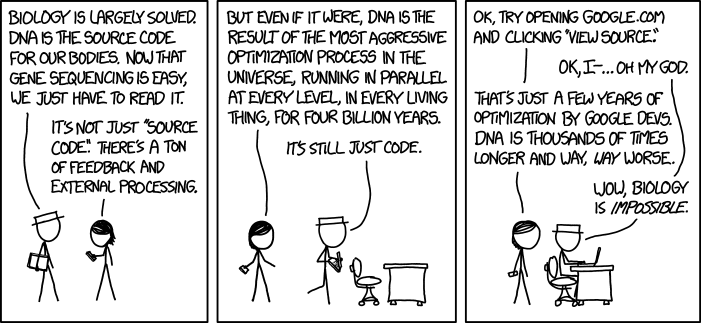
I initially tw**ted this with the comment “Truth. Biology is impossible”, because the cartoon emphasises that the information DNA contains is way more complicated than most non-biologists imagine. It’s a classic mistake of physical scientists (especially mathematicians and physicists) to think that biology obeys the same kind of rigorous lawfulness of those subjects – when they study biology they soon realise that living things are far more complicated than anything in physics or maths. As Martin Rees, the Astronomer Royal, put it: an insect is more complex than a star.
But thinking about it, I think although Randall (the author) has his heart in the right place, he’s still suffering from some the classic physical scientist’s assumptions (he used to be a NASA roboticist). Leaving aside the issue of the conditionality of gene expression (that may be what is meant by ‘feedback and external processing’ in the first panel), the female character explains that ‘DNA is the result of the most aggressive optimisation process in the universe’. It is the comparison of the 3.8 billion years of ‘optimisation’ of DNA with the few years of Google optimisation that makes the character in the hat conclude that ‘biology is impossible’.
This isn’t right. DNA is not subject to ‘the most aggressive optimisation process in the universe’. Our genes are not perfectly adapted and beautifully designed. They are a horrible, historical mess. That is partly what distinguishes biology from physics and maths – it is the outcome of historical processes – evolution and natural selection* – which leave their past traces in the genome.
For reasons we don’t understand, many eukaryotic genes (that is, genes in organisms with a nucleus – so all multicellular organisms and some single-celled forms, too) are sometimes split up, interspersed by apparently meaningless sequences, called ‘introns’. Although the average intron is only 40 bases long, one of the introns in the human dystrophin gene is more than 300,000 bases long! In some rare cases, the intron of one gene can even contain a completely separate, protein-encoding gene.
This isn’t the result of ‘optimisation’: it’s due to the fact that, as François Jacob put it, evolution does not design, it tinkers. It fiddles around with stuff to hand, and as long as it works, that’s all that matters.
We know that only 5% of the human genome encodes proteins (when Francis Crick was working on the meaning of the genetic code in the 1950s, he assumed that’s all that a gene would ever do). We now know that another 5-10% is regulatory DNA, which produces RNA that regulates the activity of other genes. As to the remaining 85% – around 2.7 billion base pairs – it appears mainly to be ‘junk’, which has no apparent function – if it were deleted, it would not affect the fitness of the organism at all.
There’s been a lot of argument about this, in particular since the ENCODE project suggested that virtually every bit of our DNA seemed to produce some kind of chemical reaction in a cell, which they argued meant that it was functional. But when scientists synthesised genuinely random bits of DNA, they found that they, too, could produce a reaction. If biochemical activity is produced by much of our DNA, it is indistinguishable from random noise.
I wrote about this in my book Life’s Greatest Secret:
Different species can have substantial differences in the size of their genomes, which do not seem to be related to anything in their ecology or degree of apparent physiological complexity. (…) This problem is called the ‘C-value paradox’ or ‘C-value enigma’ – ‘C’ is the amount of DNA in a genome. Some of these differences may be due to a well-known phenomenon: chunks of genomes can be duplicated during evolution, particularly in plants, which can double their genome size in one generation when chro- mosome duplication goes slightly awry. Because of factors such as duplication, the variation in genomic size that we see between species resists any overall functional explanation. This is highlighted by what is known jocularly as the onion test: the onion genome contains around 16 billion base pairs, or five times that of a human.
Another example – viruses can insert themselves into our DNA, using our cells to reproduce themselves. Sometimes they end up getting stuck, and are copied over and over. So, for example, the remnants of these invasive viral sequences make up an astonishing 45 per cent of the human genome, with one element, known as Alu, leaving genetic traces that make up around 10 per cent of your DNA.
That isn’t optimisation – it’s a millenia-long history of infection!
In some cases, these viral remnants can actually be repurposed by natural selection – tinkering with a vengeance – and such viral sequences are now thought to be at the origin of one of our most vital organs – the placenta.
On a final note, in some cases, within this amazing noise, there are also astonishing examples of complexity which do indeed appear to be the result of optimisation – and they would boggle the mind of anyone, not just a cocky computer scientist in a hat. In Drosophila there is a gene called Dscam, which is involved in neuronal development and has four clusters of exons (bits of the gene that are expressed – hence exon – in contrast to the apparently inert introns).
Each of these exons can be read by the cell in twelve, forty-eight, thirty-three or two alternative ways. As a result, the single stretch of DNA that we call Dscam can encode 38,016 different proteins. (For the moment, this is the record number of alternative proteins produced by a single gene. I suspect there are many even more extreme examples.)
In other words, DNA is even more complicated than Randall imagines – it is historical, messy, undesigned. And when occasionally it is optimised, the degree of complexity is mind-boggling. Biology is not quite impossible, it is just incredibly difficult!
__________________________________________________________________
* These are not the same thing! Evolution is a change in the frequency of a particular allele, or form, or a gene. Many alleles – different DNA sequences – produce no change in any character and are therefore selectively neutral. They can change their frequency without natural selection being involved.
Similarly, natural selection simply means the differential survival of different forms of organism in a species. If the differences in form have no genetic basis then natural selection will not lead to a change in allele frequencies, and therefore evolution. For example, if there was natural selection against pink flamingos, which get their colour from their environment, not their genes, then this would not lead to the evolution of a new form in the population (unless there is a genetic character for tending to go and eat only food that enables them to turn pink).
Darwin’s genius was to realise that evolution by natural selection was a way for adaptations to appear. To get this to occur you need variability in a population, for that variability to have some genetic basis and for it to lead to differential survival. With that combination of three factors, and sufficient time, you end up with the amazing variety of life we have on the planet.

“For example, if there was natural selection against pink flamingos, which get their colour from their environment, not their genes, then this would not lead to the evolution of a new form in the population (unless there is a genetic character for tending to go and eat only food that enables them to turn pink).”
I don’t think that’s right. The flamingos don’t just automatically turn pink because they eat certain food. There are evolved pathways that cause the pink color, using chemicals in the food as precursors.
If pink color led to fewer offspring, forms with disabled or altered color-synthesis pathways would eventually take over the population, producing new color forms.
Continuing with minor quibbles, if there is selection AGAINST pink flamingos, the selection would be FOR flamingos that (for genetic reasons)tended to AVOID “food that enables them to turn pink.”
Also, as Lou Jost says, and alternatively, relevant color-synthesis pathways could be disabled or altered, either in general or just for feathers.
Minor quibble. You said,
“…the female character explains that ‘DNA is the result of the most aggressive optimisation process in the universe’. It is the comparison of the 3.8 billion years of ‘optimisation’ of DNA with the few years of Google optimisation that makes the character in the hat conclude that ‘biology is impossible’.
This isn’t right. DNA is not subject to ‘the most aggressive optimisation.’ Our genes are not perfectly adapted and beautifully designed.”
The XKCD doesn’t imply that DNA is beautifully designed. Randall Munroe is likely alluding to the fact that the ‘optimization’ of the source code has rendered it utterly uninterpretable and incomprehensibly complex. Just like the DNA. In fact, you can see the source code for google.com by visiting the page, right clicking anywhere and choosing the option to view source.
I was thinking the strip used the term “optimization” in a specific way. For example, the shape of a birds wing is pretty well optimized for flying. The genetic acrobatics required to create such an optimized design are, of course, anything but clean and simple. They take the path of least resistance only at the point that some chemical bond or splice takes place – by the laws of chemistry and physics. Otherwise it’s just a matter of what mutation happens to occur and when. So maybe “optimization” can make sense when considering different levels of construction – from a mutation to the birds wing.
“evolution does not design, it tinkers. It fiddles around with stuff to hand, and as long as it works, that’s all that matters.”
Matthew, I need to introduce you to many of my fellow programmers 😀
Yeah I was going to say something similar. Matthew’s comment “Our genes are not perfectly adapted and beautifully designed. They are a horrible, historical mess” makes the analogy to human programming better, not worse. 🙂
Definitively! This was the crux of Randall’s joke if I am not mistaken.
I agree. On both counts!
What are examples of programming that a biologist would understand that are cases of horrible, historical messes? I am always looking for examples of palimpsests and other historical relics.
I would guess that the entire etymology of computer programming languages is a result of historical contingency (i.e. a horrible, historical mess) as much as it is a result of optimization or rational choice. The reason Java forms the basis of so many internet-based languages is because that’s what was included in the earliest version of Netscape Navigator, which captured the market at the time. And the reason there are so many Visual Basic type programming languages is because Basic is what ran on the first generation of IBM personal computers. Geez, I know labs that were programming their nuclear physics detector setups in Fortran in the 1990s, and that is a language invented for use with punch cards.
Now computer programming languages will probably always require a more formal and rigorous syntax than natural language, but IMO the specific formal syntaxes that we used today are more due to the vagaries of human history than they are any sort of rational choice of the best options.
For that matter, why the frak do we even bother with www? Http vs. Https? That’s four redundant and therefore worthless letters out of five, the equivalent of 80% “junk DNA” in one of the most common and most recent human-built computer syntaxes. What sense does it make? None. Why do we have it? History.
What about the classic example of the, “QWERTY” keyboard? We’re stuck with it, now, even though it was originally designed to slow the typist down.
You are thinking of Javascript, not Java.
By the way, the four letters of http are not redundant because that part of the URL (called the “scheme”) has many other options besides “http” and “https”. e.g. file, smb, ftp, ssh, ftps, sftp, smtp etc etc etc.
Here’s a simple one, which is analogous to pseudogenes. It often happens that people write programs and then add or change stuff. Often these additions and changes leave blocks of code “stranded”, by eliminating the lines which called that block of code.
Many programmers don’t care enough about these unused blocks to go search them out and remove them manually. Why bother, if the benefit of removing them is small compared to the time that would need to be spent looking for them?
A little bit of time invested up front in housecleaning can pay immeasurable dividends in future maintenance. Carefully-written, well-commented code is easy to update. Spaghetti code typically needs to be taken out behind the barn and shot.
b&
The fundamental thing to realize about any computer system/program/OS/whatever with a long history is that its design is a result of a long series of decisions, each of which made sense to a particular person at a particular time under a particular set of constraints — but not the same person at the same time under the same constraints. The more things have changed over the course of development, the less overall sense the result will make.
Here’s an example I remember from college (I was studying computer science). David Parnas gave a talk about reimplementing the flight control software for the navy’s A-7E aircraft. At the time, the A-7 was over a decade old, and had gone through numerous modifications over that time, and as a result the flight control software had evolved into a complete mess. Parnas said he’d heard that there was one person on the team that maintained the system who understood its overall control flow, but that person was on vacation when he met the team so he couldn’t confirm their existence.
There’s one example he gave that stood out to me: at some point, one of the aircrafts’s options (a weapon system? I don’t remember) had been discontinued, so it could be removed from the relevant table of options. Except it couldn’t be removed, because all the code that referred to that table was full of hard-coded addresses, and changing the table layout would’ve meant tracking down all the interdependent code that assumed particular locations and changing them… it was easier to just leave a blank space at that point in the table.
Then sometime later, they ran low on space for executable code (this was an old system, very tightly memory-constrained). But there was some space in that table that wasn’t really being used for anything… so they crammed the extra executable code in the middle of the weapons(?) table.
I remember thinking that the time thinking how utterly biological it sounded. I still think that.
BTW, while the old system was a complete mess organizationally, it worked perfectly well; after all, it had been through over a decade of debugging and optimization. Parnas said he considered his ground-up reimplementation to be a success because it worked as well as the original.
The history of computing is full of “we gotta do this this way because version 1.0 did it that way and we can’t break [other process].” Sounds a lot like biological evolution. On the other hand, programming is now trying to get away from when that happens with well defined interfaces, “future proofing” and so on, which are not available in the biological context.
As it happens one can see “voices” in programming style, if the team members are allowed to vary things a little – and often even if they aren’t.
I always thought the funniest examples were the obviously incorrect programing nonsense that hides behind if tests that can never be true.
As to the remaining 85% – around 2.7 billion base pairs – it appears mainly to be ‘junk’, which has no apparent function – if it were deleted, it would not affect the fitness of the organism at all.
Odd. I could have sworn I read somewhere in Dawkins’ books that apparently junk DNA does have a passive function: while not synthesizing or regulating the rest of the genome directly, its removal would throw the sequence out of balance and so is necessary for normal protein synthesis and RNA modulation to occur. Was what I read speculation, then?
Each of these exons can be read by the cell in twelve, forty-eight, thirty-three or two alternative ways. As a result, the single stretch of DNA that we call Dscam can encode 38,016 different proteins. (For the moment, this is the record number of alternative proteins produced by a single gene. I suspect there are many even more extreme examples.)
Firstly, does it count towards the number of proteins attributable to these exons if not all of them are, in fact, used, or are used in conjunction with exons elsewhere on the genome? Or is it really the exact same exons simply read in different ways?
Secondly, how is that diversity created? If there are multiple different ways to read the same bit of code, then where are those multiple ways themselves encoded? Are they all scattered across the genome as genes that code for RNA that modulates the reading? Or is it that there are multiple RNA readers that share the protein codes among themselves?
I recollect that Dawkins used junk DNA to illustrate his point about the selfish gene.
Hmm. Apparently Dawkins changed his story in light of the ENCODE project results, to the delight of his detractors.
That he had the courage and honesty to admit he got it wrong makes him more worthy of admiration imo. It’s a shame many of his detractors don’t share that quality.
The point his detractors are delighted with is that his change of position is wrong. ENCODE did not show that most of the human genome is functional; it’s still mostly junk, and Dawkins (or anyone else) should realize that.
I agree. But the detractors do make one valid point, I think. Dawkins claimed the existence of what he thought was non-functional DNA demonstrated the theory of natural selection and then turned around, when some of it was found to be functional afterall, and said *that* demonstrated natural selection. The correct statement, I believe, is that both functional and non-functional DNA are consistent with natural selection. That is, DNA functionality is not a test of natural selection. I will be happy to be shown I am wrong about this.
Thanks for the explanation. 🙂
As usual, I got it a wrong when I tried to comment on science-y stuff, but my point about his character stands I think.
I hope this is correct.
Prokaryotes have little junk DNA.
This is due to
1) large populations (neutral and “nearly neutral” mutations less likely to be fixed) and
2)intense darwinian competition (very slight deleterious effect of carrying junk is significant).
Larger multi cellular organisms acquire a lot of junk DNA because of
1) small populations (neutral and “nearly neutral” mutations can more easily become fixed – therefore creating more junk)
2) the effect of carrying junk DNA isn’t significant in darwinian terms (compared to other selection pressures).
My impression is that superfluous DNA – not all of it junk genes – have a lot of secondary functions known or suggested, such as regulating gene expression and cell size et cetera. But nothing that explains the C-value enigma.
I’d love to hear from a biologist on this.
“The production of alternatively spliced mRNAs is regulated by a system of trans-acting proteins that bind to cis-acting sites on the primary transcript itself. Such proteins include splicing activators that promote the usage of a particular splice site, and splicing repressors that reduce the usage of a particular site. Mechanisms of alternative splicing are highly variable, and new examples are constantly being found, particularly through the use of high-throughput techniques. Researchers hope to fully elucidate the regulatory systems involved in splicing, so that alternative splicing products from a given gene under particular conditions could be predicted by a “splicing code”.[3][4]”
[ https://en.wikipedia.org/wiki/Alternative_splicing ]
Thank you. I cringed when I read that word optimization. Life is a freaking mess, with no end of redundancy.
The same with optimization of computers (at least those that I know).
Natural selection does ‘aim’ at optimizing the phenotype such that fitness (at the appropriate level) is maximized. Or have I got it wrong?
But what a mess. I give you the goshawk.
http://tinyurl.com/lmy2k3t
“Optimization” doesn’t have to mean “perfection.” To me it means, “making the best of what you have to work with.” So far DNA hasn’t evolved an “edit function” so there’s nothing that can be optimized in that regard.
Nearly as bad as politics.
Of course, Deepak Chopra and countless other pseudo-scientists think that the guy in the hat in that cartoon is a true representation of a typical geneticist. They have taken on the task of protecting the public from those who practice such scientism.
“DNA is not subject to ‘the most aggressive optimisation process in the universe’. Our genes are not perfectly adapted”
Couldn’t we just say that the optimization is very short-sighted?
I think the first part of the joke is that the result of the process of people optimizing software, as it actually occurs, is considerably more messy than the goal the term “optimisation” describes.
The more optimisation the more bloated, convoluted and hard to understand the code becomes.
Like with biological systems, source code can be optimized for several distinct purposes. The google page code is presumably optimized for download time to be minimal and perhaps also for speed of execution/rendering. On the other hand, some other code (perhaps not in a web page) would be optimized to use little memory, be readable, be more secure, etc. (Which isn’t to say that G. doesn’t have these in mind, but that they are often trade offs.)
sub
I took it to mean the most aggressive optimization process, since there is little if any optimization elsewhere in nature.
For myself, I prefer Randall’s funny description.
I think this points to a fundamental misunderstanding between what constitutes an optimization process that tries to optimize in some sense – here by hill climbing a sufficiently dense fitness measure – and optimization/optima – here hitting a fitness peak – which can be a result. With all respect for contingency, genetic drift, superfluous DNA, et cetera.
I know only of T Gregory Ryan’s version. If I remember correctly it is the tetraploidy of some onions that are nearly identical with others of less ploidy that is the illustration of the C-value enigma.
Can’t “optimization” be understood as a process of improvement without necessarily attaining some optimum?
Tout est pour le mieux dans le meilleur des mondes possibles
You can certainly analyze putative adaptations for functional performance in nature and measure how far they fall short of maximizing fitness (i.e., evaluate their approximation to optimality). It is not metaphysics.
Great post. Regardless of the complexity or remarkable messiness of DNA, life is still defined by how it functions in a physical world and responds to stimuli.
At present life has only responded to life on earth. Imagine an alien race much like X-men with powers to controls minds by manipulating chemical processes in the brain at a distance. Our collective DNA would be outclassed by this species. Our only hope, in the short term, would be to mate with them in the hopes that we acquire some of their DNA. And that species too may be outclassed by another alien species capable of manipulated energy, and so forth.
Ultimately, the species that wins is the one whose DNA figures out how to manipulate nature the best. And so far, the competition has not been all that difficult to overcome.
When I was just 2 years old, I suffered from an autoimmune disease called Guillain-Barré Syndrome, or GBS. The immune system mistakenly attacks the myelin, which is basically an insulator for the nerves. That means signals from the brain can barely or cannot be conducted. The result was that my legs became paralyzed for a few weeks. I don’t remember much of it, but my oldest memory is that I started to walk again. Only 1 in every 100.000 people develop GBS per year. Lucky me. This is hardly optimisation.
On a theological note: GBS is also why I find it difficult to believe our bodies are designed. Who would make a machine that tries to paralyze itself after just 2 years? If our bodies are designed, I’ve got some issues to settle with the designer about mine. And ofcourse I don’t believe that the disabled can be cured by miracles. I was disabled and I was cured by doctors.
And I thought having pimples and glasses made the designer argument a crock.
DNA = random variations. Some good, some bad.
I go many further. Why can’t I play piano like Rachmaninoff? Why can’t I memorize like a grand master? Why can’t I have the oration skills of Hitchens? Why can’t I swim like Phelps?
I am designed for crap, especially since my desires far exceed my capabilities.
The theist’s retort would be along the lines: “If you train hard enough, I’m sure you can achieve a lot.” I prefer to use my personal experience as an example, because it sticks to the point of God the Designer; autoimmune diseases can’t be blamed on something external like bacteria.
Jerry: ” Although the average intron is only 40 bases long,”
I believe this is incorrect. Both mean and median intron size is much longer – see Table 2 in the following paper: http://mbe.oxfordjournals.org/content/23/12/2392/T2.expansion.html
The article was written by Matthew Cobb.
You could both use some clarification. Average intron, but averaged over what genome or genomes? As you can see from the table, the average intron lengths in the four species cited are quite different, and the average of the four species would be still another.
Intron length increases with the overall genome size, and mean and median intron size will differ greatly across species, so it is true that “average intron size” is meaningless unless talking about a single specific genome. However in a large majority of plant and animal genomes introns will be much longer than 40bp on average. Only some fungi with very compact genomes and rare oddballs among animals (such as Oikopleura) will have short introns.
Do you have a reference for intron length increasing with genome size? It makes sense, since the same sorts of things that add to intron length would also tend to add to the lengths of other sequences. But I’d like to see a plot.
Here is the paper describing the relationship in amniote vertebrates:
http://gbe.oxfordjournals.org/content/4/10/1033.full
Other processes can complicate the relationship, such as whole genome duplications which are more common in some other lineages.
Evolution is not optimizing the genome to be optimal in the sense of optimally clear, readable, cleanly “designed”, etc. It is optimizing it to perform particular tasks well – whatever natural selection is pushing it to do better. Programmers who know what they’re doing warn about the dangers of “premature optimization” – whenever you take a nice, beautiful piece of code and try to push that code toward doing a particular task really well (really quickly, really efficiently), the code rapidly becomes completely disgusting. Unreadable, incomprehensible, unmaintainable, brittle. Worse still is if you optimize a piece of code for one purpose, and then later you realize you want it to be optimal for some other task instead, so you try to add further optimizations to the already optimized code, toward the new target; often this renders the code completely useless and you’re better off just throwing it out and starting over from scratch. This is what natural selection does, over countless millennia: the target of optimization is constantly shifting, as the changing environment changes the selective pressures that are being applied. Except that evolution never gets to throw out the source code and start over from scratch; it has to work with what it’s got. So the code just gets ever more disgusting and unmaintainable and brittle. That’s the point of the comic. So I think Munroe has it exactly right.
She’s not kidding about loading google.com and viewing the source. It’s amazing how complex such a simple-looking web page can be. It has a size of 424,243 bytes.
The term everybody is groping towards but nobody’s yet used, is, “spaghetti code.”
Today’s task at work is a minor example of such. Some code originally whipped up as a quick-and-dirty means to do something simple that was never going to be used again…got pulled out again later and tweaked a bit to do something slightly different…and again and again and again…and now it’s, logically, doing nothing more than a very straightforward listing of vendors who haven’t submitted an invoice in a year or more and their account’s current balance, but along the way it’s needlessly calculating all sorts of other sums and doing other extraneous stuff. Each layer gets piled on and interwoven with everything already there, until the result resembles a pile of spaghetti more than anything else.
In computer programming, we sometimes get the luxury of taking the sword to the Gordian Knot, which is my job for the day. When I’m done, it’ll be intelligently designed…but, when I started, it was most definitely evolved.
Cheers,
b&
Don’t talk to me about spaghetti code!
But years back, I wrote a little one-page hack in a very primitive basic called ‘asic’, to search a friend’s parts catalog. It sort of grewed from there. It soon became unmanageable in asic, so it got translated into Quickbasic. When it outgrew that (I *think* from experiment Quickbasic had a 64k limit on source code – at compile time I was having to strip all the comments out of my source code, and then combine four separate 64k chunks of code plus several libraries, thanks so much Bill Gates) I switched to an open-source and very powerful, though now obsolescent, Basic called Xbasic. Originally written to control telescopes, I believe. Where it still is. About 300k of source code. I do try to prune the spaghetti from time to time.
It *should* have been written in perl or Dbase 3 or similar from the start, but I didn’t know either of those languages (and dbase 3 cost $$$).
Oh, and I’m an engineer not a programmer…
cr
Yep. That’s how you get spaghetti code. Hire an engineer. 😉
I didn’t know Sergio Leone wrote code.
The good, the bad, and the ugly. Mostly the ugly.
Every gene tells a story, every genome is a hoarder and to call it optimisation is folly as I am lead to believe there is no end goal, just adaptations, traits to make a living by.
Unraveling billions of years sounds like a labour of love. Fascinating and I for one am blown away with the complexity that is life and grateful I’m alive now to take a glimpse.. thanks for the post.
Hey Matt,
Optimization in code doesn’t mean what you think it means. From wikipedia:
“In computer science, program optimization or software optimization is the process of modifying a software system to make some aspect of it work more efficiently or use fewer resources.”
Optimization of programs is the step where they become most illegible, unchangeable, and unscalable. It’s the period during which you are tweaking for the outside factors at the severe detriment of your coding cleanliness. Optimiization is usually among the last steps, as it makes altering your code very difficult.
Natural selection is an extremely aggressive optimization process. Conducting this optimization while still making major changes is very complex.
There’s even a saying:
“Premature optimization is the root of all evil.”
Or as I like to say, “There’s no sense in running fast if one cannot run correct.”
(The trick is knowing what is correct, since “exactly right” may not be optimal for other reasons, like being too slow or even computationally unsolvable.)
“…correctly.”
Researchers just found the gene responsible for mistakenly thinking we’ve found the gene for specific things. It’s the region between the start and the end of every chromosome, plus a few segments in out mitochondria.
– mouseover text (what hell is that called) in xkcd
By the way, did anyone try xkcd’s suggestion and view the page source at google.com? Randall chose his example carefully.
The page source at xkcd is fairly comprehensible, the source of this page of WEIT is not too bad (but with a chunk of C code near the bottom, or is that php?), but the page source of Google is indeed solid alphabet soup.
cr
I call it, “mouseover text.”
I’ve just remembered, ‘Alt tags’. Which is more concise but probably more obscure to non-geeks.
cr
Genomes are very unintelligently designed, aren’t they?
Very nice article. Thank you.
Agree! Matthew makes everything so clear and interesting.
There seems to be a lot of consensus here that Randall Munroe has got the biological side of the analogy wrong. I’m not an expert in biology but I am an expert in programming and I can tell you that he has the programming side of the analogy wrong too.
Contrary to what the menu option says, when you click “view source” to see what’s behind Google’s search page, you are not seeing the source code. What you see is called minified Javascript. It is the result of taking the original, possibly elegant, possibly well designed code and stripping it of everything that makes it legible to humans so that it can be sent across the Internet more quickly.
Given that it’s Google, the original source may not even have been Javascript, it may have been written in some other language and compiled into Javascript so that it will run in the browser.
Anyway, what you see when you “view source” is not the analogy of DNA, it’s more like the analogy of what you see when you cut a hole into nature’s most elegant, sleekest and agile “creation” (the cat) and find out it is full of wobbly, jellylike, stringy, and slimy bits.
True, ‘view source’ in the browser (or email, so long as you’re not using MS Outlook) doesn’t necessarily represent the original source code, what it shows is what was received down the line from the Internet, before your browser interpreted it into the page on your screen.
When the term ‘view source’ was first included in browsers, that probably was the original HTML source you were seeing. It was also possible (and still is) to create your own simple webpage (i.e. one without pop-ups, forms, cookies and all the other gremlins that lurk in ‘modern’ pages) just by ‘borrowing’ someone else’s page source and substituting your own text and pictures with a plain text editor. No fancy webpage creation software required.
You can usually tell pages so created because they are actually readable in a text editor, each HTML tag was put there for a purpose, whereas many software-created pages are full of useless bandwidth-gobbling junk HTML that has been attached to every line of text just repeating the tags from the previous line. (Very similar to junk DNA in that respect).
cr
So the stuff that works is the intelligent design and the junk is the scribbled notes and failed experiments, and the Designer decided, oh wtf, let’s keep everything?